Compare Read and Write files time
When we are dealing with large datasets, and we need to write many csv files or when the csv filethat we hand to read is huge, then the speed of the read and write command is important. We will compare the required time to write and read files of the following cases:
- base package
- data.table
- readr
Compare the Write times
We will work with a csv file of 1M rows and 10 columns which is approximately 180MB. Let’s create the sample data frame and write it to the hard disk. We will generate 10M observations from the Normal Distribution
library(data.table)
library(readr)
library(microbenchmark)
library(ggplot2)
# create a 1M X 10 data frame
my_df<-data.frame(matrix(rnorm(1000000*10), 1000000,10))
# base
system.time({ write.csv(my_df, "base.csv", row.names=FALSE) })
# data.table
system.time({ fwrite(my_df, "datatable.csv") })
# readr
system.time({ write_csv(my_df, "readr.csv") })

As we can see from the elapsed time, the fwrite from the data.table is ~70 times faster than the base package and ~7times faster than the readr
Compare the Read Times
Let’s compare also the read times using the microbenchmark package.
tm <- microbenchmark(read.csv("datatable.csv"),
fread("datatable.csv"),
read_csv("datatable.csv"),
times = 10L
)
tm
autoplot(tm)

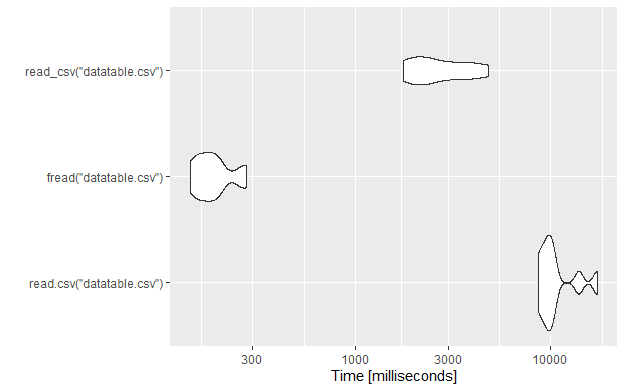
As we can see, again the fread from the data.table package is around 40 times faster than the base package and 8.5 times faster than the read_csv from the readr package.
Conclusion
If you want to read and write files fastly then you should choose the data.table package.



4 thoughts on “The fastest way to Read and Write files in R”
Have you tried this with smaller data frames (say 10000 entries)? Is it still worth moving from read.csv to fread in this case?
I know, potential gain is small, but sometimes I have to load dozens of such small tables, so every (micro)second matters…
Excelent!, Thank you very much, so simple and so smashing efficient!
Thank you