Introduction
We can analyze different scientific studies that address the same question by applying a meta-analysis. The assumption is that every individual study contains some degree of error. For example, a study could be the mortality rate of two treatments for a specific disease. The goal is to obtain pooled summary estimates from individual studies by taking into consideration the heterogeneity among individual studies. The aggregated data from the individual studies leads to higher statistical power.
Procedure
- Define the research question
- Define inclusion/exclusion criteria for individual studies screened
- Search literature
- Select eligible studies
- Collect data
- Aggregate findings across studies and obtain pooled estimates of effect size
- Evaluate heterogeneity of included studies
- Conduct sensitivity and subgroup analyses
Statistical Models
- Pool effect sizes (of individual studies) into one overall effect
- Two types of statistical models to combine data, Fixed Effect Model and Random Effects Model
- Frequentist vs Bayesian approach
Example of Meta Analysis in R
We will provide an example of Meta Analysis in R using the meta library. Let’s start.
library(meta)
data("Fleiss1993cont")
head(Fleiss1993cont)
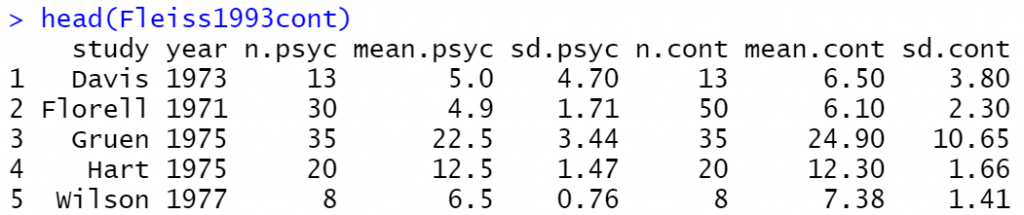
We will work with the Fleiss1993cont dataset where:
| study | study label |
| year | year of publication |
| n.psyc | number of observations in psychotherapy group |
| mean.psyc | estimated mean in psychotherapy group |
| sd.psyc | standard deviation in psychotherapy group |
| n.cont | number of observations in control group |
| mean.cont | estimated mean in control group |
| sd.cont | standard deviation in control group |
# meta-analysis with continuout outcome
# comb.fixed/comb.random: indicator whether a fix/random effect mata-analysis to be conducted.
# sm: Three different types of summary measures to choose,standardized mean difference (SMD),mean difference (MD), ratio of means (ROM)
res.flesiss = metacont(n.psyc, mean.psyc, sd.psyc,
n.cont, mean.cont, sd.cont,
comb.fixed = T, comb.random = T, studlab = study,
data = Fleiss1993cont, sm = "SMD")
res.flesiss
Output
SMD 95%-CI %W(fixed) %W(random)
Davis -0.3399 [-1.1152; 0.4354] 11.5 11.5
Florell -0.5659 [-1.0274; -0.1044] 32.6 32.6
Gruen -0.2999 [-0.7712; 0.1714] 31.2 31.2
Hart 0.1250 [-0.4954; 0.7455] 18.0 18.0
Wilson -0.7346 [-1.7575; 0.2883] 6.6 6.6
Number of studies combined: k = 5
Number of observations: o = 232
SMD 95%-CI z p-value
Fixed effect model -0.3434 [-0.6068; -0.0800] -2.56 0.0106
Random effects model -0.3434 [-0.6068; -0.0800] -2.56 0.0106
Quantifying heterogeneity:
tau^2 = 0 [0.0000; 0.6936]; tau = 0 [0.0000; 0.8328]
I^2 = 0.0% [0.0%; 79.2%]; H = 1.00 [1.00; 2.19]
Test of heterogeneity:
Q d.f. p-value
3.68 4 0.4515
Details on meta-analytical method:
- Inverse variance method
- DerSimonian-Laird estimator for tau^2
- Jackson method for confidence interval of tau^2 and tau
- Hedges' g (bias corrected standardised mean difference)Forest
forest(res.flesiss, leftcols = c('studlab'))
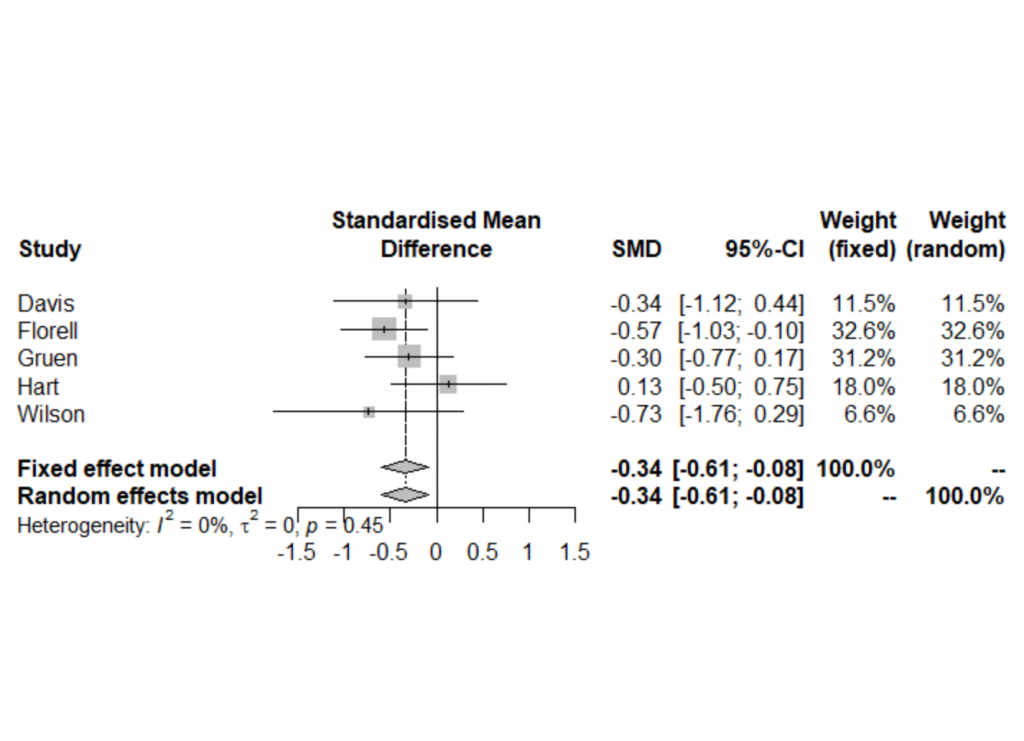
- According to the pooled results of meta-analysis, both fixed and random effects models yield a significant benefit of the intervention group against the control group (for the days of hospital stay, the lower, the better).
- The p-value =0.45 for the Cochran’s Q test, indicating no heterogeneity.
Funnel Plot
funnel(res.flesiss)
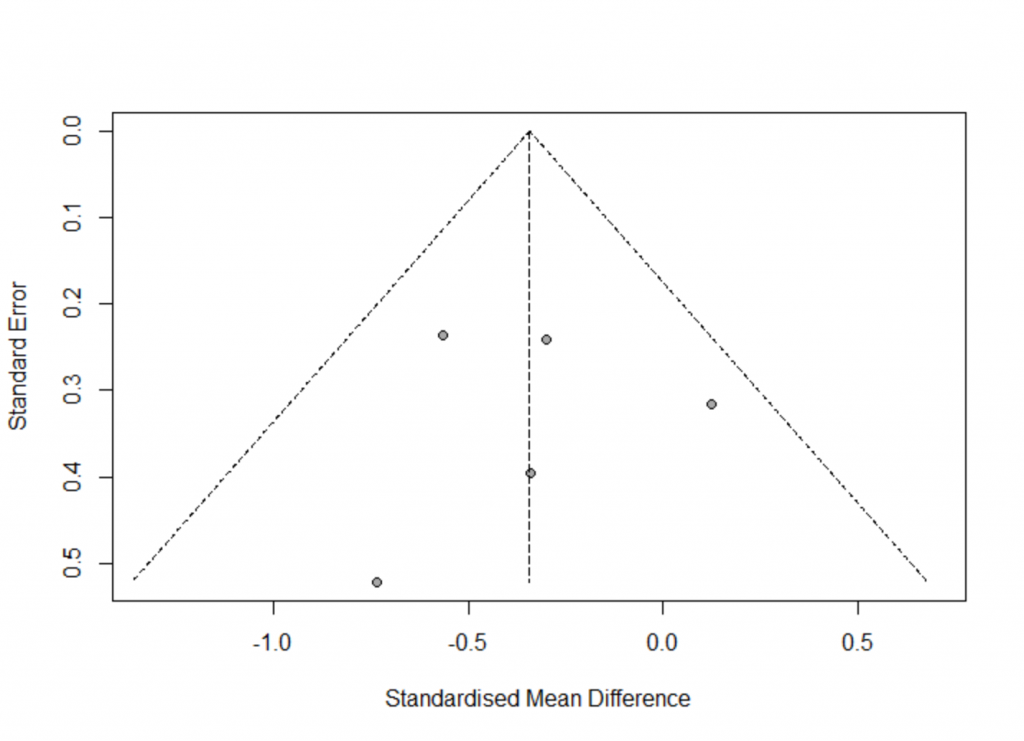
- metabias: Test for funnel plot asymmetry, based on rank correlation or linear regression method.
- Use Egger’s test to check publication bias, can take string ‘Egger’ or ‘linreg’.
metabias(res.flesiss, method.bias = 'linreg', k.min = 5, plotit = T)
Output
Linear regression test of funnel plot asymmetry
Test result: t = -0.04, df = 3, p-value = 0.9730
Sample estimates:
bias se.bias intercept se.intercept
-0.0668 1.8154 -0.3241 0.5455
Details:
- multiplicative residual heterogeneity variance (tau^2 = 1.2251)
- predictor: standard error
- weight: inverse variance
- reference: Egger et al. (1997), BMJ
The p-value is 0.973 which implies no publication bias. However, this meta-analysis contains k=5 studies. Egger’s test may lack the statistical power to detect bias when the number of studies is small (i.e., k<10).
Binary case
At this point, we will provide an example by taking into consideration a binary case.
load("binarydata.RData")
binarydata
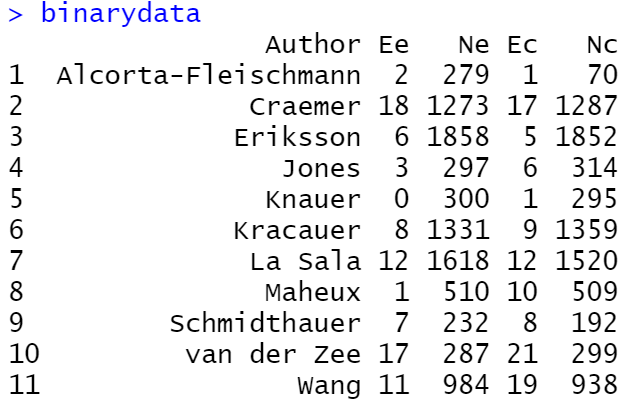
- Author: This signifies the column for the study label (i.e., the first author)
- Ee: Number of events in the experimental treatment arm
- Ne: Number of participants in the experimental treatment arm
- Ec: Number of events in the control arm
- Nc: Number of participants in the control arm
The Analysis:
- Use metabin to do the calculation.
- As we want to have a pooled effect for binary data, we have to choose another summary measure now. We can choose from “OR” (Odds Ratio), “RR” (Risk Ratio), or RD (Risk Difference), among other things.
- method: indicating which method is to be used for pooling of studies.
m.bin <- metabin(Ee,Ne,Ec,Nc,
data = binarydata,
studlab = paste(Author),
comb.fixed = T,comb.random = T,
method = 'MH',sm = "RR") # Mantel Haenszel weighting
Output
RR 95%-CI %W(fixed) %W(random)
Alcorta-Fleischmann 0.5018 [0.0462; 5.4551] 1.4 1.4
Craemer 1.0705 [0.5542; 2.0676] 15.2 18.9
Eriksson 1.1961 [0.3657; 3.9124] 4.5 5.8
Jones 0.5286 [0.1334; 2.0945] 5.3 4.3
Knauer 0.3278 [0.0134; 8.0140] 1.4 0.8
Kracauer 0.9076 [0.3512; 2.3453] 8.0 9.1
La Sala 0.9394 [0.4233; 2.0847] 11.2 12.9
Maheux 0.0998 [0.0128; 0.7768] 9.0 1.9
Schmidthauer 0.7241 [0.2674; 1.9609] 7.9 8.3
van der Zee 0.8434 [0.4543; 1.5656] 18.5 21.4
Wang 0.5519 [0.2641; 1.1534] 17.5 15.1
Number of studies combined: k = 11
Number of observations: o = 17604
Number of events: e = 194
RR 95%-CI z p-value
Fixed effect model 0.7536 [0.5696; 0.9972] -1.98 0.0478
Random effects model 0.7885 [0.5922; 1.0499] -1.63 0.1038
Quantifying heterogeneity:
tau^2 = 0; tau = 0; I^2 = 0.0% [0.0%; 60.2%]; H = 1.00 [1.00; 1.59]
Test of heterogeneity:
Q d.f. p-value
7.29 10 0.6976
Details on meta-analytical method:
- Mantel-Haenszel method
- DerSimonian-Laird estimator for tau^2
- Mantel-Haenszel estimator used in calculation of Q and tau^2 (like RevMan 5)
- Continuity correction of 0.5 in studies with zero cell frequenciesForest
forest(m.bin, leftcols = c('studlab'))

funnel(m.bin) metabias(m.bin, method.bias = 'linreg', plotit = T)
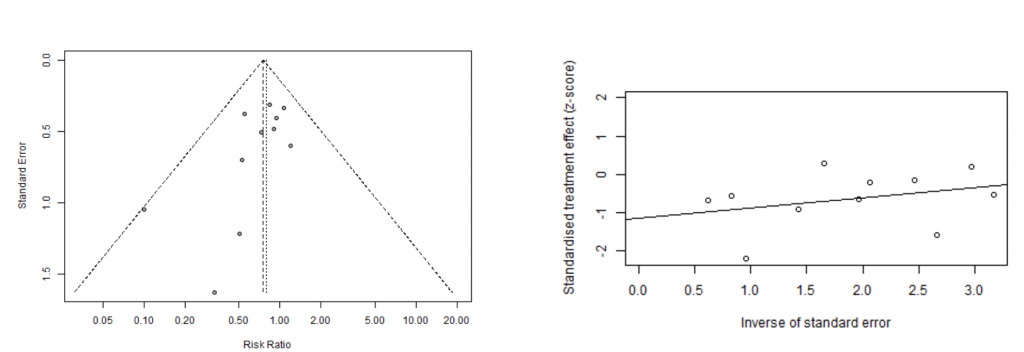
Linear regression test of funnel plot asymmetry
Test result: t = -2.05, df = 9, p-value = 0.0701
Sample estimates:
bias se.bias intercept se.intercept
-1.1286 0.5493 0.2623 0.2661
Details:
- multiplicative residual heterogeneity variance (tau^2 = 0.5443)
- predictor: standard error
- weight: inverse variance
- reference: Egger et al. (1997), BMJLimitations
The limitation of Meta Analysis is that it can only conduct pariwise comparisions, and cannot include multi-arm trials.
References
[1] Dr. Na Zhao



1 thought on “Meta Analysis in R”
Thank you very much for this article.
One question. Is it possible to do a meta-analysis if I am missing the binary data and only have the Hazard ratios and confidence intervals? Thank you