We have provided an example of K-means clustering and now we will provide an example of Hierarchical Clustering. We will work with the famous Iris Dataset.
import pandas as pd import numpy as np import matplotlib.pyplot as plt import seaborn as sns %matplotlib inline from sklearn import datasets iris = datasets.load_iris() df=pd.DataFrame(iris['data']) print(df.head())
0 1 2 3
0 5.1 3.5 1.4 0.2
1 4.9 3.0 1.4 0.2
2 4.7 3.2 1.3 0.2
3 4.6 3.1 1.5 0.2
4 5.0 3.6 1.4 0.2Let’s see the number of targets that the Iris dataset has and their frequency:
np.unique(iris.target,return_counts=True)
(array([0, 1, 2]), array([50, 50, 50], dtype=int64))As we can see there are three targets of 50 observations each. If we want to see the names of the target:
iris.target_names
array(['setosa', 'versicolor', 'virginica'], dtype='<U10')Data Preparation for Cluster Analysis
When we apply Cluster Analysis we need to scale our data. There are many different approaches like standardizing or normalizing the values etc. Also, we can whiten the values which is a process of rescaling data to a standard deviation of 1:
\(x_{new} = x/std\_dev(x)\)
Let’s scaled the iris dataset.
# Import the whiten function from scipy.cluster.vq import whiten scaled_data = whiten(df.to_numpy())
Let’s check if the variance of every feature is close to 1 now:
pd.DataFrame(scaled_data).describe()
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 7.080243 | 7.037882 | 2.135951 | 1.578709 |
| std | 1.003350 | 1.003350 | 1.003350 | 1.003350 |
| min | 5.210218 | 4.603935 | 0.568374 | 0.131632 |
| 25% | 6.179561 | 6.445509 | 0.909399 | 0.394897 |
| 50% | 7.027736 | 6.905903 | 2.472428 | 1.711218 |
| 75% | 7.754744 | 7.596493 | 2.898709 | 2.369379 |
| max | 9.572262 | 10.128658 | 3.921782 | 3.290805 |
Creat the Distance Matrix based on linkage
Look at the documentation of the `linkage` function to see the available methods and metrics.
# Import the fcluster and linkage functions from scipy.cluster.hierarchy import fcluster, linkage # Use the linkage() function distance_matrix = linkage(scaled_data, method = 'ward', metric = 'euclidean')
How many Clusters – Introduction to dendrograms
Dendrograms help in showing progressions as clusters are merged. It is a branching diagram that demonstrates how each cluster is composed by branching out into its child nodes.
# Import the dendrogram function from scipy.cluster.hierarchy import dendrogram # Create a dendrogram dn = dendrogram(distance_matrix) # Display the dendogram plt.show()
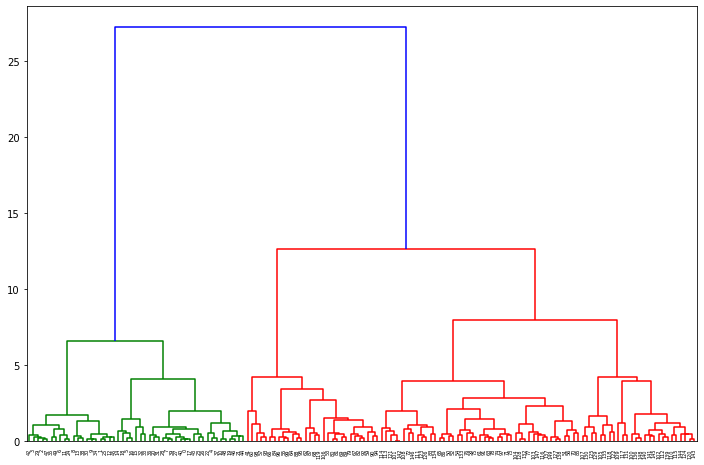
From the dendrogram we can realize that a good candidate for the number of Clusters is 3 and that 2 clusters are closer (the red ones) compared to the green one.
Run the Hierarchical Clustering
# Assign cluster labels df['cluster_labels'] = fcluster(distance_matrix, 3, criterion='maxclust')
Notice that we can define clusters based on the linkage distance by changing the criterion to distance in the fcluster function!
Hierarchical vs Actual for n_clusters=3
df['target'] = iris.target
fig, axes = plt.subplots(1, 2, figsize=(16,8))
axes[0].scatter(df[0], df[1], c=df['target'])
axes[1].scatter(df[0], df[1], c=df['cluster_labels'], cmap=plt.cm.Set1)
axes[0].set_title('Actual', fontsize=18)
axes[1].set_title('Hierarchical', fontsize=18)